- Study of novel CRISPR-Cas systems and their biotechnological applications
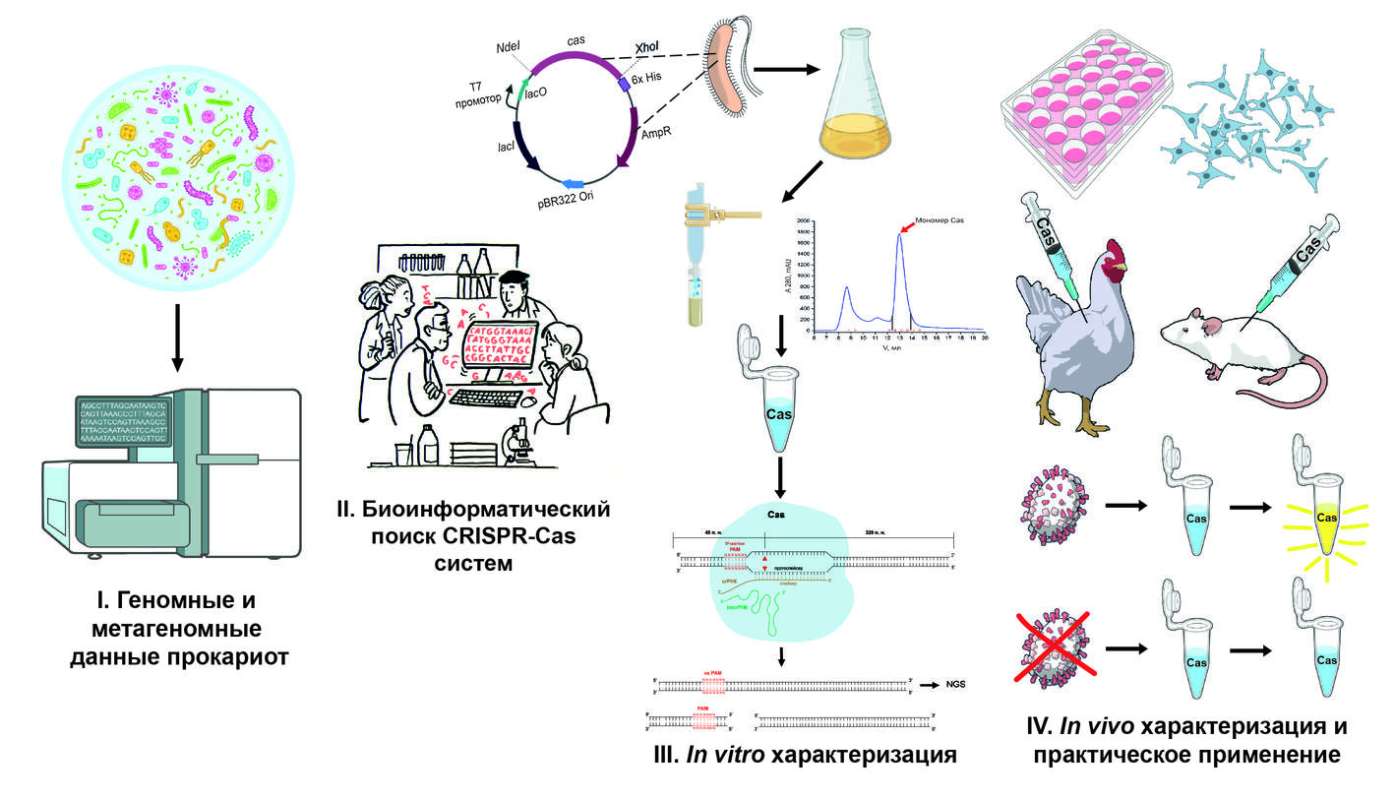
CRISPR-Cas – is a system found in bacteria and archaea that serves as their natural immunity against viral infections. The CRISPR locus contains repetitive DNA regions and spacers — fragments of viral DNA that cells have encountered before. When a virus attacks, the information from the CRISPR locus is converted into guide RNAs, which bind to Cas proteins. These proteins cut the viral DNA, rendering the virus harmless.
The main scientific discovery is that this natural mechanism can be used to edit the genomes of other organisms. By manipulating guide RNA, scientists can force Cas proteins to target specific regions of the genome. This use of CRISPR-Cas systems has become a powerful tool in molecular biology.Our group is engaged in the search for and study of new CRISPR-Cas systems, discovering them from a wide variety of sources. We work with carefully studied genomes and metagenomes from large databases, as well as with unique samples provided by our collaborators. First, using computer analysis (in silico) we identify potential CRISPR-Cas candidates. Once a suitable system is identified, we move on to in vitro laboratory testing– we check whether the selected candidate is capable of cutting the DNA fragments we propose in a targeted manner. If the system shows activity, we proceed in two ways, depending on the goals. In the first case, we test the system in vivo, testing its activity in human cell cultures. This allows us to determine its effectiveness and potential application for genome editing. In the second case, we adapt the CRISPR-Cas system for use in diagnostics – It becomes the basis of a method that helps to quickly and accurately determine the presence of specific pathogens (bacteria and viruses) in the samples being studied.
Our goal is not just to study new CRISPR-Cas systems, but to translate them into active use in research and practical applications.
Main publications:
1. Vasilyeva A.A., Alyukas S.A., Selkova P.A., Arseniev A.N., Chernova V.E., Musharova O.S., Klimuk E.I., Khodorkovsky M.A., Severinov K.V. CRISPR-Cas type II nucleases: search algorithm and in vitro characterization // Molecular Biology. – 2023. – Т. 57. – №3. – C. 546–560. doi: 10.31857/S0026898423030163
2. Vasileva A, Selkova P, Arseniev A, Abramova M, Shcheglova N, Musharova O, Mizgirev I, Artamonova T, Khodorkovskii M, Severinov K, Fedorova I. Characterization of CoCas9 nuclease from Capnocytophaga ochracea. RNA Biol. 2023 Jan;20(1):750-759. doi: 10.1080/15476286.2023.2256578
3. Fedorova I, Vasileva A, Selkova P, Abramova M, Arseniev A, Pobegalov G, Kazalov M, Musharova O, Goryanin I, Artamonova D, Zyubko T, Shmakov S, Artamonova T, Khodorkovskii M, Severinov K. PpCas9 from Pasteurella pneumotropica – a compact Type II-C Cas9 ortholog active in human cells. Nucleic Acids Res. 2020 Dec 2;48(21):12297-12309. doi: 10.1093/nar/gkaa998
4. Fedorova I, Arseniev A, Selkova P, Pobegalov G, Goryanin I, Vasileva A, Musharova O, Abramova M, Kazalov M, Zyubko T, Artamonova T, Artamonova D, Shmakov S, Khodorkovskii M, Severinov K. DNA targeting by Clostridium cellulolyticum CRISPR-Cas9 Type II-C system. Nucleic Acids Res. 2020 Feb 28;48(4):2026-2034. doi: 10.1093/nar/gkz1225
Key patents:
1. RU2820345C1 «A Nucleic Acid Detection Tool Based on ScCas12a Protein from Bacteria Sedimentisphaera cyanobacteriorum» (2024)
2. RU2804422C1 «A system for editing the genomic DNA of a eukaryotic cell based on the nucleotide sequence encoding a protein SuCas9NLS» (2023)
3. RU2778156C1 «DNA cutting tool based on Cas9 protein from bacteria Capnocytophaga ochracea» (2022)
4. RU2712497C1 «DNA cutting agent based on Cas9 protein from biotechnologically important bacteria Clostridium cellulolyticum» (2020)







- Development of a universal diagnostic platform based on CRISPR-Cas technologies for the detection of infectious diseases of various origins
Description of research
Today, one of the main challenges facing humanity is the emergence of new strains of viruses and bacteria. To prevent the spread of infections, it is important to quickly and accurately diagnose them at the early stages of the disease. Most of the diagnostic methods used today are expensive, require specialized equipment, and take a relatively long time to obtain diagnostic results.
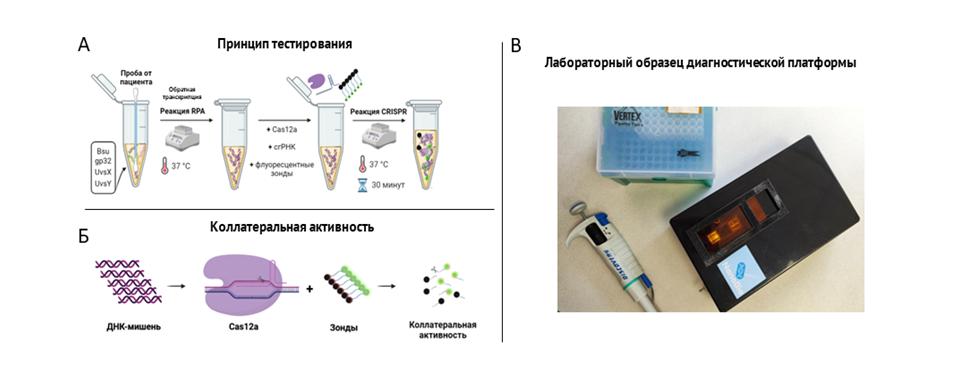
Developed in NIK "NanoBio" A diagnostic platform based on the reaction of isothermal amplification (RPA) and the collateral activity of the CRISPR-Cas system has a sensitivity comparable to RT-PCR (103 copies), at the same time, the reaction time is relatively short (30-40 minutes) and does not require additional laboratory equipment, since the entire testing process takes place at a fixed temperature (37-39°C). This opens up broad prospects for its use outside the laboratory without loss of diagnostic accuracy.










- Study of the development of cell infection by giant bacteriophages
Description of research
Bacteriophages are bacterial viruses that are a molecule of nucleic acid packed in a protein shell outside a bacterial cell. When a bacteriophage encounters a bacterium in which it can reproduce, it transfers its genetic information into the bacterium, which initiates a program for the formation of new phage particles.Our main object of study is the giant bacteriophage phiKZ and related phages. During the 60 minutes that pass from the moment of injection of phiKZ phage DNA to the release of newly synthesized phage particles, all processes inside the cell are radically rebuilt and the morphology of the bacterium is significantly changed: the bacterial nucleoid changes its position, giving way to a central spherical compartment resembling the nucleus of a eukaryotic cell. This compartment is covered with a protein shell, inside which the phage DNA and some of the proteins involved in transcription and replication are packed, in addition, it is held in the center of the cell by tubulin-like phage proteins. For transcription of its genome, the phiKZ phage exclusively uses its DNA-dependent RNA polymerases, of which it has two.
Our group studies the processes occurring inside the cell against the background of infection with the phiKZ phage and the mechanisms responsible for their implementation.
Main publications:
1. Danilova Y.A.; Belousova V.V.; Moiseenko A.V.; Vishnyakov I.E.; Yakunina M.V.; Sokolova O.S. Maturation of Pseudo-Nucleus Compartment in P. aeruginosa, Infected with Giant phiKZ Phage, Viruses 2020, 12, 1197
2. Orekhova, M., Koreshova, A., Artamonova, T., Khodorkovskii, M., Yakunina, M. The study of the phiKZ phage non-canonical non-virion RNA polymerase, Biochemical and Biophysical Research Communications, 2019, doi: 10.1016/j.bbrc.2019.02.132
3. Lavysh D, Sokolova M, Minakhin L, Yakunina M, Artamonova T, Kozyavkin S, Makarova KS, Koonin EV, Severinov K. The genome of AR9, a giant transducing Bacillus phage encoding two multisubunit RNA polymerases, Journal of Virology – 2016. – Vol. 495. – P. 185-196.
4. Yakunina M, Artamonova T, Borukhov S, Kira S. Makarova, Severinov K, Minakhin, L. A non-canonical multisubunit RNA polymerase encoded by a giant bacteriophage, Nucleic Acids Research. - 2015. - Vol. 43. – P. 10411-10420.
5. Natàlia de Martín Garrido, Orekhova M., Yuen Ting Emilie Lai Wan Loong, Anna Litvinova, Kailash Ramlaul, Tatyana Artamonova, Alexei S Melnikov, Pavel Serdobintsev, Christopher H S Aylett, Maria Yakunina. Structure of the bacteriophage PhiKZ non-virion RNA polymerase, Nucleic Acids Research, 2021, Volume 49, Volume 49, Issue 13, P. 7732–7739, doi: 10.1093/nar/gkab539





- Single-molecule studies of DNA-protein interactions
Description of research
The group focuses on studies of DNA-protein interactions using the optical capture method implemented on the Laser Tweezers setup. The combination of the optical capture method with a microfluidic laminar flow system allows tracking changes in the mechanical properties of individual DNA molecules during interaction with DNA-binding proteins. Using the optical capture method, our group studies the properties of nucleoprotein complexes formed by the bacterial protein RecA (E. coli, D. radiodurans) on duplex and single-stranded (single-stranded) DNA molecules. In bacteria, RecA in the form of a nucleoprotein filament formed on single-stranded DNA is involved in several key processes at once: reparation of severe DNA damage, transfer of antibiotic resistance genes, and initiation of the SOS response. It is considered known that, depending on the type of bound nucleotide cofactor, the RecA filament on single-stranded DNA can be in an active or inactive conformation. To maintain the active conformation of the filament, the presence of free ATP in the medium is necessary. The ATPase activity of RecA potentially leads to the local emergence of areas of inactive conformation in the structure of the filament, which generally has an active conformation. Our group conducts studies of conformational rearrangements and states of the RecA filament, as well as the regulation of RecA activity by protein cofactors.
The group also studies bacterial transcription and its regulation at the single-molecule level in real time using the acoustic force spectroscopy method. We are the first in the world to successfully apply this single-molecule approach to study transcription processes. The developed technique allowed us to identify the nature of inhibition of new lasso-peptides acinetodine and klebsidin. At the moment, we are preparing a publication that presents a unique method for synergistic inhibition of RNA polymerase by the antibiotic streptolydigin together with the transcription factors GreA and GreB. Our research in this area not only expands our understanding of the molecular mechanisms of bacterial transcription, but may also have important practical applications in the development of new antimicrobial drugs.
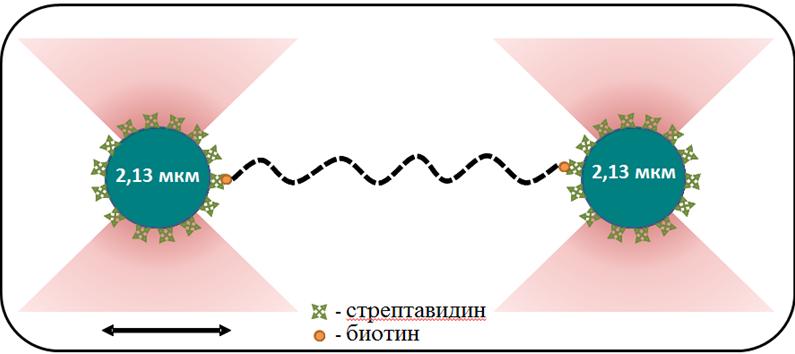
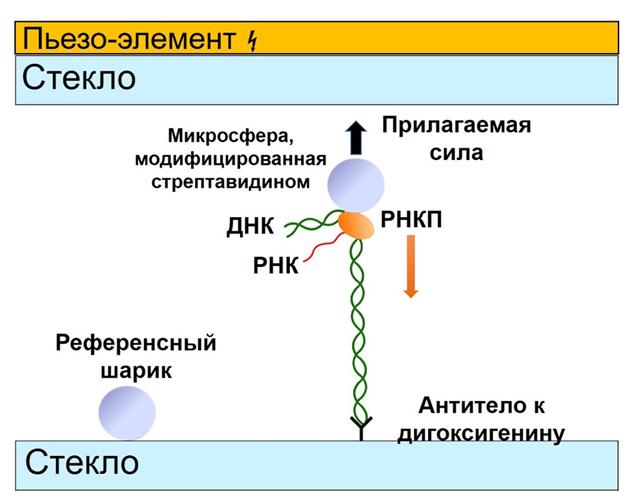
Main publications:
1. Alekseev A., Pobegalov G., Morozova N., Vedyaykin A., Cherevatenko G., Yakimov A., Baitin D., Khodorkovskii M., A new insight into RecA filament regulation by RecX from the analysis of conformation-specific interactions, eLife, 2022, V. 11, e78409
2. Alekseev, A., Serdakov, M., Pobegalov, G., Yakimov, A., Bakhlanova, I., Baitin, D., Khodorkovskii, M. Single‐molecule analysis reveals two distinct states of the compressed RecA filament on single‐stranded DNA. FEBS letters, 2020, 594(21), 3464-3476.
3. Alekseev, A., Cherevatenko, G., Serdakov, M., Pobegalov, G., Yakimov, A., Bakhlanova, I., Baitin, D., Khodorkovskii, M. Single-Molecule Insights into ATP-Dependent Conformational Dynamics of Nucleoprotein Filaments of Deinococcus radiodurans RecA. International journal of molecular sciences, 2020, 21(19), 7389.
4. Alekseev A., Morozova N., Vedyaykin A., Yakimov A., Khodorkovskii M., Pobegalov G., Single-molecule characterization of compressed RecA nucleoprotein filaments, Biochemical and Biophysical Research Communications, 2022, V. 614, pp. 29-33
5. Yakimov, A., Pobegalov, G., Bakhlanova, I., Khodorkovskii, M., Petukhov, M., Baitin, D. Blocking the RecA activity and SOS-response in bacteria with a short α-helical peptide. Nucleic acids research, 2017, 45(16), 9788-9796.
6. Pobegalov, G, Cherevatenko, G., Alekseev, A., Sabantsev, A., Kovaleva, O., Vedyaykin, A., Morozova, N., Baitin, D., Khodorkovskii, M. Deinococcus radiodurans RecA nucleoprotein filaments characterized at the single-molecule level with optical tweezers. Biochemical and biophysical research communications, 2015, 466(3), 426-430.
7. Metelev M, Arseniev A, Bushin LB, Kuznedelov K, Artamonova TO, Kondratenko R, Khodorkovskii M, Seyedsayamdost MR, Severinov K. Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae. ACS Chem Biol. 2017 Mar 17;12(3):814-824




- Study of molecular mechanisms of restriction-modification systems and other bacterial defense systems against viruses at the level of single bacterial cells
Description of research
In living nature, bacteria are constantly exposed to infection by viruses called bacteriophages. Such viruses are capable of destroying entire bacterial populations extremely effectively. To protect themselves from viral infection, bacteria have developed many molecular mechanisms that act on completely different parts of the bacteriophage at different stages of its development in the cell.
One of the most common bacterial defense systems is the restriction-modification system. Such systems operate through the activity of restriction endonuclease, which introduces a double-strand break into specific recognition sites on DNA, and methyltransferase activity, which methylates the bacterial genome, protecting it from degradation by the restriction endonuclease. Despite the high level of protection provided by these systems, in some cases the DNA of the infecting bacteriophage escapes degradation and undergoes modification. As a result, modified viral progeny arises, completely resistant to the action of the restriction-modification system.
We study the influence of variation in the expression of restriction-modification system genes on the efficiency of protection of single bacteria from the virus. We also study the mechanisms of operation of other bacterial protection systems from the virus at the level of single bacteria.
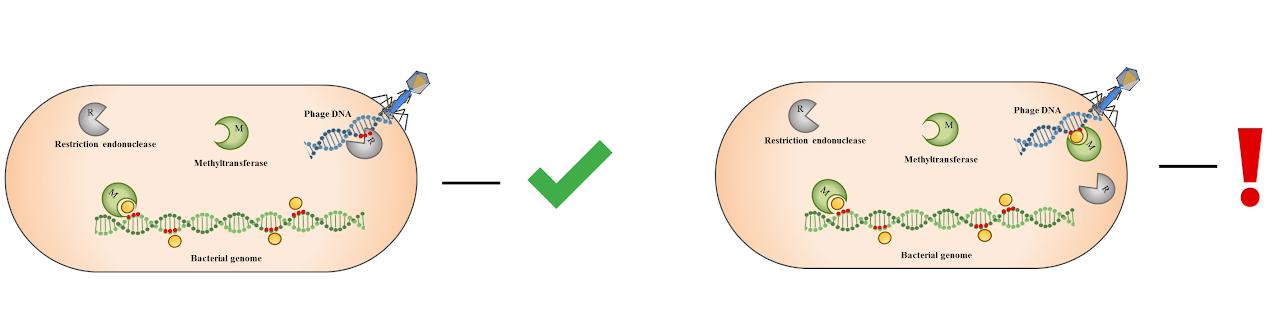
Main publications:
1. Morozova N, Sabantsev A, Bogdanova E, Fedorova Y, Maikova A, Vedyaykin A, et al. Temporal dynamics of methyltransferase and restriction endonuclease accumulation in individual cells after introducing a restriction-modification system. Nucleic Acids Research, 2016, 44(2), 790-800. DOI: 10.1093/nar/gkv1490.
2. A. Strotskaya, E. Savitskaya, A. Metlitskaya, N. Morozova, K. Datsenko, E. Semenova, K. Severinov. The action of Escherichia coli CRISPR–Cas system on lytic bacteriophages with different lifestyles and development strategies. Nucleic Acids Research, 2017, 45(4), 1946-1957. DOI: 10.1093/nar/gkx042.
3. B. Wilcox, I. Osterman, M. Serebryakova, D. Lukyanov, E. Komarova, B. Gollan, N. Morozova, Yu. Wolf, K. Makarova, S. Helaine, P. Sergiev, S. Dubiley, S. Borukhov, K. Severinov. Escherichia coli ItaT is a type II toxin that inhibits translation by acetylating isoleucyl-tRNAIle. Nucleic Acids Research, 2018, 46(15), 7873–7885. DOI: 10.1093/nar/gky560.
4. Smirnov, S.V., Morozova, N.E., Khodorkovskii, M.A., Severinov, K.V. Fluorescence microscopy study of the effect of Esp1396I restriction-modification system proteins concentrations on protection against lambda phage. Journal of Physics: Conference Series, 2018, 1135(1), 012016. DOI: 10.1088/1742-6596/1135/1/012016.
5. J. Gordeeva, N. Morozova, N. Sierro, A. Isaev, T. Sinkunas, K. Tsvetkova, M. Matlashov, L. Truncaitė, R.D. Morgan, N.V. Ivanov, V. Siksnys, L. Zeng, K. Severinov. BREX system of Escherichia coli distinguishes self from non-self by methylation of a specific DNA site. Nucleic Acids Research, 2019, 47(1), 253–265. DOI: doi.org/10.1093/nar/gky1125.
6. Antonova, D.A., Morozova, N.E., Shiryaeva, A.A., Khodorkovskii, M.A. Regulation of type II restriction-modification system Esp1396I. Journal of Physics: Conference Series, 2019, 1400(3), 033024. DOI: 10.1088/1742-6596/1400/3/033024.
7. Znobishcheva, E.A., Morozova, N.E., Khodorkovskii, M.A. Fluorescent labeling of bacteriophage T7 by CRISPR-Cas9. Journal of Physics: Conference Series, 2019; 1400(3), 033005. DOI: 10.1088/1742-6596/1400/3/033005.



- Study of SMC complexes and bacterial division proteins
Description of research
Cytoskeleton is a protein «cell frame». For a long time, cytoskeleton proteins were considered to belong only to eukaryotic cells - indeed, why do bacteria need a cytoskeleton if they are covered with a rigid cell wall? However, over the past 30 years, homologues of all the main eukaryotic cytoskeleton proteins – actin, tubulin and intermediate filaments - have been discovered in bacteria. These proteins in bacteria participate in various vital processes, including maintaining the shape of the cell, segregation of DNA molecules, and cell division. One of the most important proteins of the bacterial cytoskeleton is FtsZ – a key division protein that is a homologue of tubulin. How exactly does the FtsZ protein participate in bacterial division? Much is still unknown, but in well-studied bacterial species, such as Escherichia coli, The role of the FtsZ protein is generally clear. FtsZ polymerizes in the cell and forms a Z-ring, which is a framework, a kind of «scaffolding» for other division proteins. It is these proteins that are believed to perform the main work of building a partition between future daughter cells and some other functions, and FtsZ only directs these proteins, ensuring their coordinated work. However, one should not assume that everything is already known about bacterial division. What is the exact structure of the Z-ring? Do FtsZ polymers deform the cell membrane? How do bacteria divide that do not have the FtsZ protein? These and many other questions still need to be answered. Our group is studying the properties of the FtsZ protein: we visualize the structures that this protein forms in bacterial cells and in a test tube; determine which proteins it interacts with; find out how the roles and functions of the FtsZ protein differ in different types of bacteria. In addition, we aim to find out how bacteria divide that do not have the FtsZ protein. Finally, we are interested in bacterial proteins that form cytoskeleton-like structures.
Protein complexes of the SMC type (from the English Structural maintenance of chromosomes, hereinafter referred to as SMC complexes) are key participants in the spatial organization of DNA in all living organisms - in bacteria, archaea and eukaryotes. In bacteria, there are several homologues of SMC complexes that perform, at first glance, unrelated functions, but act, apparently, through very similar, conservative mechanisms. In recent years, it has been established that SMC complexes are capable of forming loops from DNA (the so-called loop extrusion), which allows us to consider them as a separate class of DNA translocases. It is interesting to note that the key property of SMC complexes – loop extrusion – has not yet been directly demonstrated for bacterial SMC complexes, unlike eukaryotic SMC complexes. The exact mechanism of extrusion has not yet been established, but several models have been proposed that require experimental confirmation. There are many unanswered questions about SMC complexes, including bacterial ones, among which, in addition to the loop extrusion mechanism, one of the most important questions is the complete composition of SMC complexes, including regulatory proteins that activate or inhibit the activity of the complexes. It is still unclear which factors control the work of SMC-like complexes - for example, ensure the coordination of their work with the cell cycle, and also allow parallel work of several types of SMC complexes in one cell. Bacteria are more simply structured, have fewer types of SMC complexes and, probably, fewer factors regulating the work of these complexes, so the study of bacterial SMC-like complexes seems to be a simpler task that can provide answers to questions that are true for SMC complexes in general. Our group is conducting a comprehensive study of several types of bacterial SMC-type complexes.
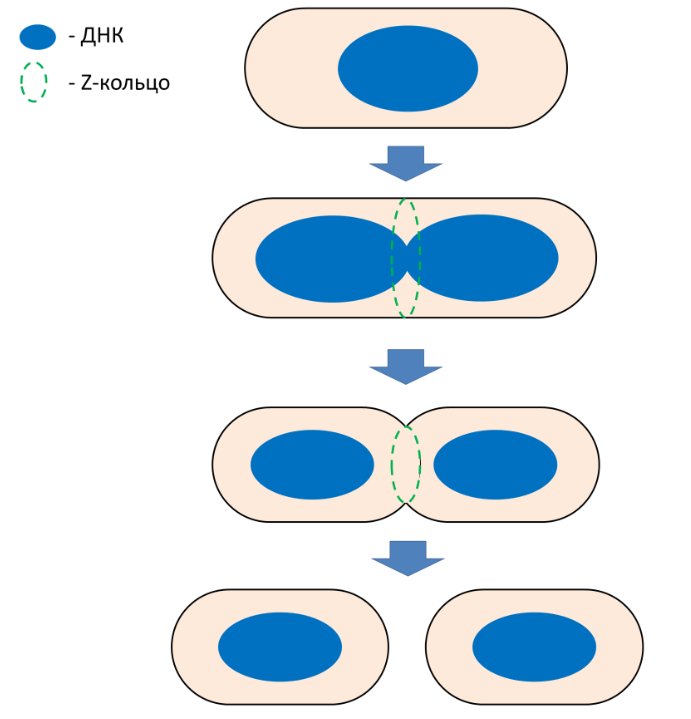
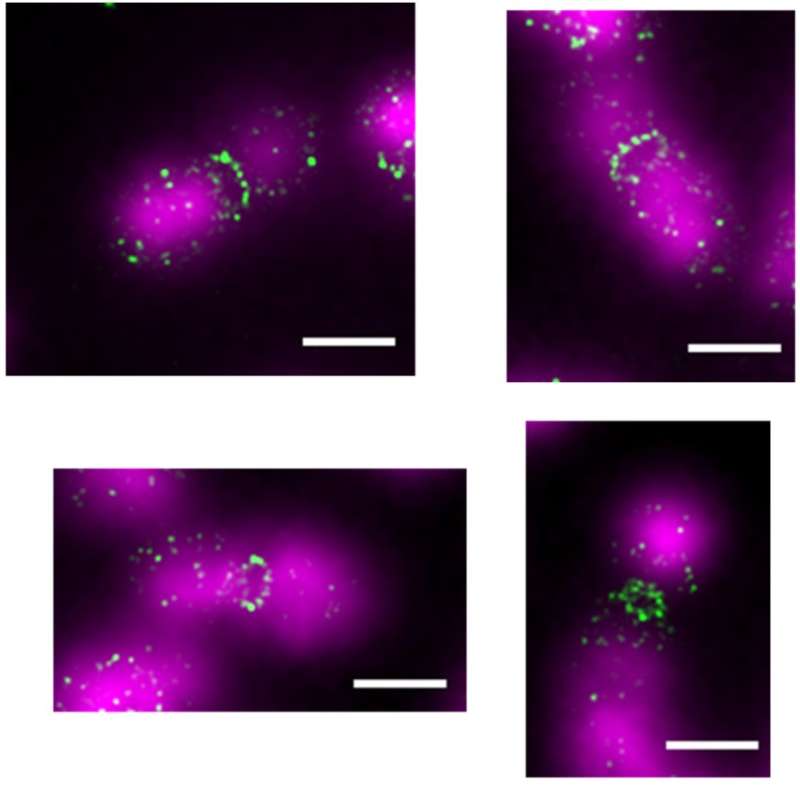
Main publications:
1. Roshektaeva V.D., Alekseev A.A., Vedyaykin A.D., Vinnik V.A., Baitin D.M., Bakhlanova I.V., Pobegalov G.E., Khodorkovskii M.A., Morozova N.E. Features of the DNA Escherichia coli RecN interaction revealed by fluorescence microscopy and single-molecule methods. // Biochemical and Biophysical Research Communications, 2024. 716: p. 150009. DOI: 10.1016/j.bbrc.2024.150009
2. Rumyantseva N.A., Golofeeva D.M., Vedyaykin A.D. SulA does not sequester FtsZ in Escherichia coli cells during the SOS response. // Biochemical and Biophysical Research Communications, 2024. 691: p. 149313. https://www.sciencedirect.com/science/article/pii/S0006291X23014079?via%3Dihub DOI: 10.1016/j.bbrc.2023.149313
3. Rumyantseva N.A., Golofeeva D.M., Shabalina A.V., Vedyaykin A.D. Direct evidence of the ability of Pseudomonas aeruginosa and E. coli SulA to dimerize. // Archives of Biochemistry and Biophysics, 2024. 751: p. 109826. https://www.sciencedirect.com/science/article/pii/S0003986123003259 DOI: 10.1016/j.abb.2023.109826
4. N. A. Rumyantseva, D. M. Golofeeva, A. A. Khasanova, A. D. Vedyaykin. Homologues of tubulin in bacteria and archaea. // Microbiology, 2024. 93. № 3. 249-266. https://link.springer.com/article/10.1134/S002626172460469X DOI: 10.1134/S002626172460469X.
5. Morozova N.E., Potyseva A.S., Vedyaikin A.D. Features of the structure and functions of bacterial SMC complexes. Cytology.2023. Т. 65, № 6 С. 522-534. https://journals.rcsi.science/0041-3771/article/view/232793 DOI: S004137712306007X.









- Study of RecN protein - SMC-like component of bacterial SOS response
Description of research
RecN is one of the factors of the SOS response, a process that occurs in a bacterial cell in response to DNA damage. The cause of such damage, in particular, may be UV radiation or the impact of various types of antibacterial agents. The result of activation of the SOS response may be the precise restoration of damaged DNA sections, as well as the adaptation of bacteria to the damaging effect, due to increased mutagenesis in bacterial cells. It is important to note that due to this, the SOS response is the main mechanism of adaptation of bacteria to the action of antibiotics.
The RecN protein is expressed and acts at an early stage of the SOS response, and according to in vivo data, is one of the key factors in the successful course of this process. It has been established that RecN belongs to the SMC proteins (Structural Maintenance of Chromosomes) - a family of ATPases involved in the dynamic regulation of the organization of the chromosome structure. SMC proteins are highly conserved among both prokaryotes and eukaryotes.



- Kinetics of ultrafast processes in molecules and clusters
Description of research
Femtosecond light pulses are used to study fast-moving processes. Such pulses are created using a femtosecond titanium-sapphire laser. Laser pulses (800 nm) are converted using a harmonic generator (400 and 266 nm) and a white light generator (supercontinuum radiation 400-1000 nm). Measurements are performed using the «excitation-probing» method using an optical delay line. The first pulse excites the object, the second pulse arrives at the object with a delay and is used to analyze the state of the object. By changing the delay, it is possible to obtain information about the processes occurring in the object under study.
Our group conducts research on fast processes in luminescent probes and inert gas clusters. It should be noted that various nonlinear optical processes, in particular multiphoton absorption, are easily observed when using femtosecond laser pulses.
Luminescent probes are widely used to measure various parameters (temperature, oxygen concentration, pH) in biological research, medicine, aerospace industry and microelectronics. A luminescent probe is a molecule (or particle) whose optical properties strongly depend on the measured value. As a rule, the intensity of luminescence or the lifetime of the excited state are measured.
One of the ways to significantly improve the properties of luminescent probes is to use them as part of molecular dyads. A molecular dyad is two chromophore centers connected by a small covalent bridge. In this type of molecular systems, after the excitation of one chromophore center, energy transfer between the chromophores is possible. The use of molecular dyads, where the chromophores have different functions and exchange energy, allows varying the properties of the systems as a whole (changing the position of absorption and/or emission bands, increasing/decreasing the measurement range, etc.). Our research is aimed at determining the mechanisms of energy transfer in molecular dyads in an electronically excited state.
Clusters of inert gases are a convenient model object for studying relaxation processes in polyatomic systems. For a number of polyatomic systems in an electronically excited state, the main relaxation mechanism is nonradiative processes. As a rule, their rate significantly exceeds the rate of radiative decay. In polyatomic molecules with relatively high binding energies between atoms, the main and often the only mechanism of nonradiative relaxation is electron-vibrational interaction. In clusters (polyatomic molecules, nanodroplets) with relatively low binding energies between atoms, additional relaxation channels appear (desorption of atoms, ionization, fragmentation). In this case, the dominant relaxation channel depends not only on the binding energy between atoms, but also on the size of the object and the excitation energy. Despite the active study of these processes in clusters, even qualitative relaxation mechanisms are not always clear today. In xenon clusters, we discovered an ICD-like process. This process is possible with simultaneous excitation of two atoms included in the cluster. The energy transfer between two adjacent excited atoms leads to ionization of the cluster. The characteristic ionization time of the xenon cluster in this process is ~ 5 ps.


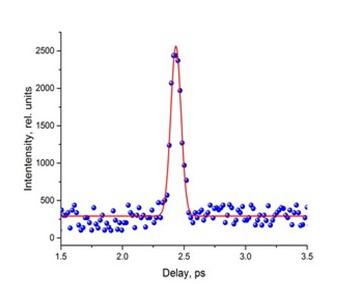

Основные публикации:
1. P.Y. Serdobintsev, A.S. Melnikov, A.A. Pastor, N.A. Timofeev, M.A. Khodorkovskiy, J. Chem. Phys., 2018, V.148, 194301
2. I. Balmaev, P. Serdobintsev, A. Melnikov, A. Pastor, M. Khodorkovskiy, Journal of Physics: Conference Series 1410 (1), 012135
3. Kisel K.S., Melnikov A.S., Grachova E.V., Hirva P., Tunik S. P., Koshevoy I.O., Chemistry–A European Journal, 2017, V.23, P.11301-11311
4. Belyaev A., Kolesnikov I., Melnikov A.S., Gurzhiy V.V., Tunik S.P., Koshevoy I.O., New Journal of Chemistry, 2019, V.43, P.13741-13750



